Hello,
I just installed CCPN V3.02 for assignment of short peptide. I faced the problem of incorrect display of the chemical shifts on the spectra calibrated in Topspin. Especially in HSQC and 1D 13C spectra. Incorrect means, that CCPN place my peak few hundreds away from where I want.
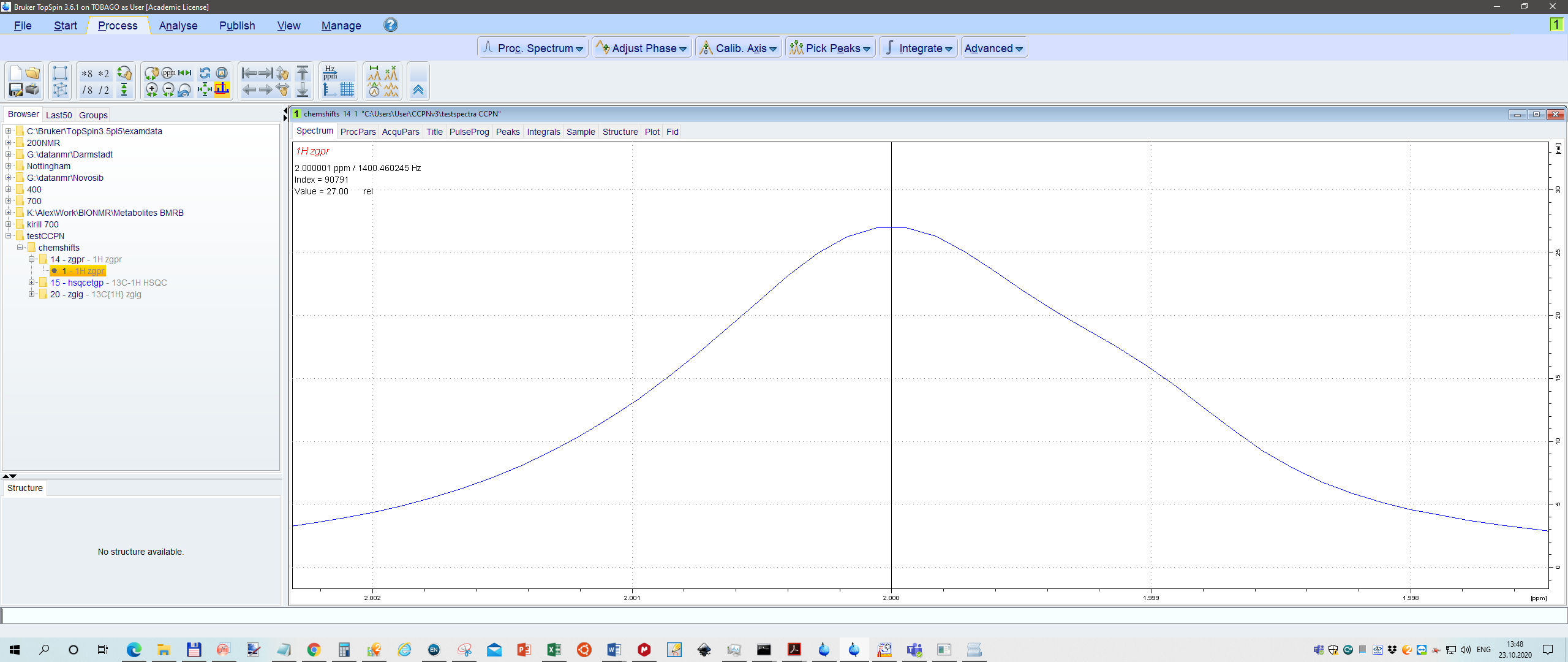
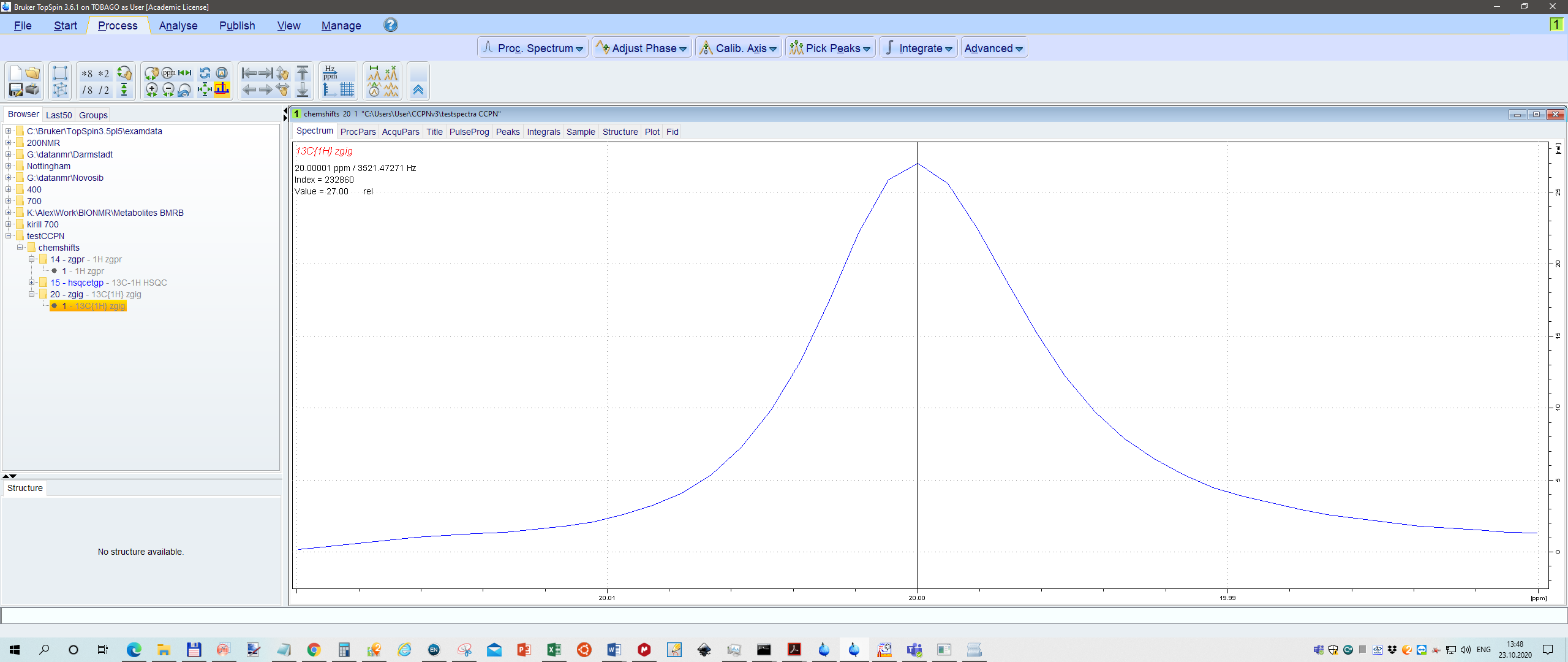
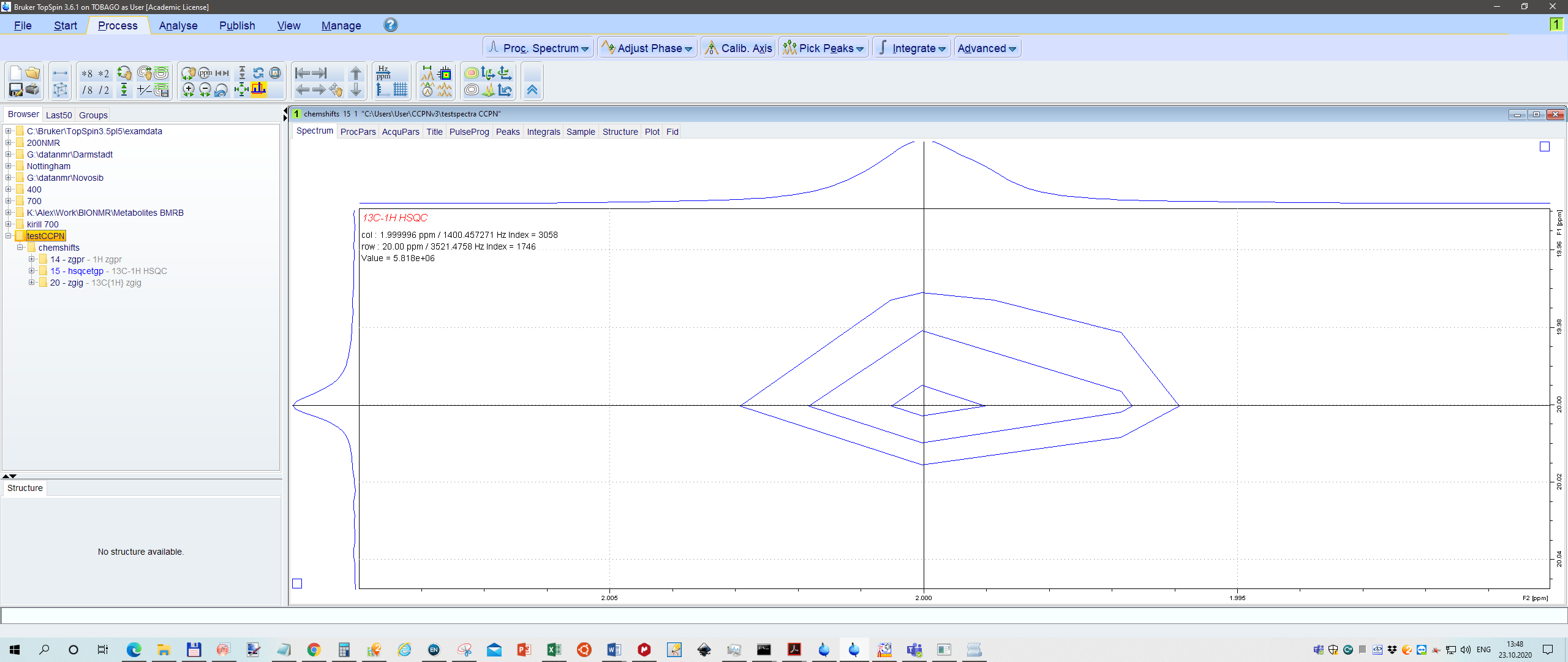
What I have done. Measured three spectra: 1H, 13C, and 13C-1H HSQC. Found strong and narrow CH3 peak (Acetate ion) and calibrated it in all three spectra using TOPSPIN (so SR is not zero anymore).
Just for fun I calibrated 1H peak to 2.000ppm, 13C peak 20.000, and also in HSQC 2.0 and 20.0 ppm, respectively.
Se screenshot
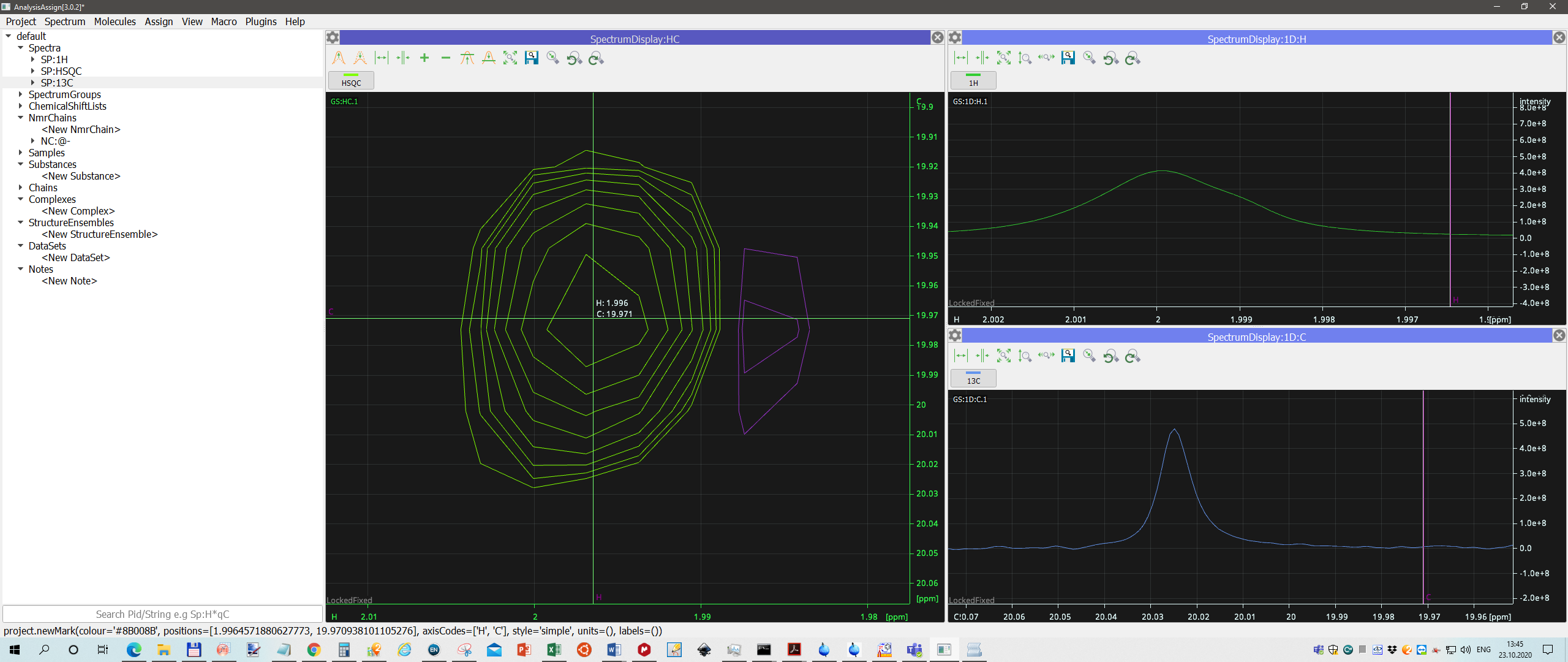
Then I load these spectra into CCPN analysis
And now I see that 1. projections are not correlated with 2D spectrum, 2. Only 1H spectrum is more or less correct, but peak is still a bit out of 2.00 ppm.
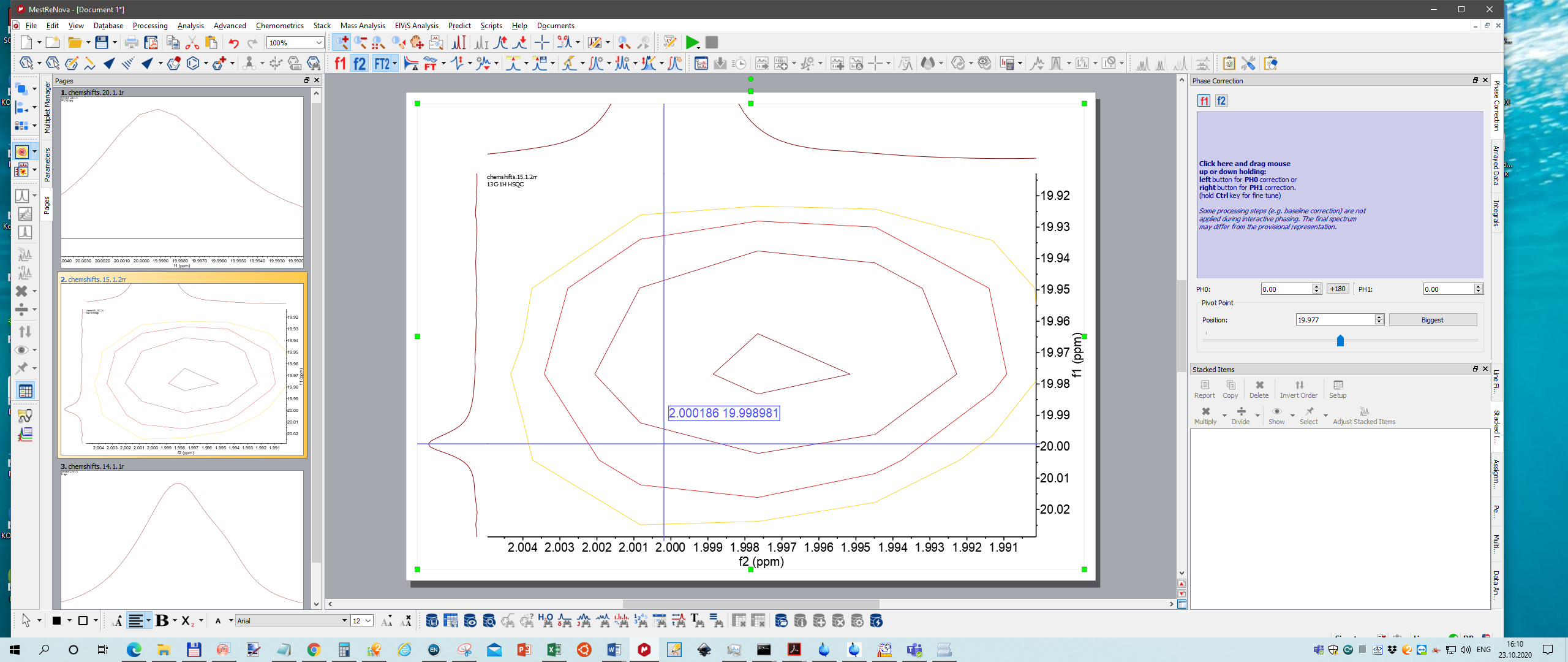
For 13C spectrum misalignment is dramatic.
see screenshot.

By the way Mnova also displays incorrectly 2D spectrum.

Please check the code for the calculation of ppm.
I know that I always can calibrate spectrum in CCPN, but I was really surprised to see that.
All the best,
Alexey
Hi Alexey,
thanks for this.
We think there is probably an issue here that Bruker data is currently being shown with an offset of 1 point (the difference in resolution/digitisation between your 1Ds and 2Ds makes this particularly obvious in your case). You can obviously change this manually in the referencing tab of spectrum properties. We`re still not quite sure where this issue is coming from (interesting that MNova has the same issue), but as we are in the process of re-writing our Bruker import functions, we`ll make sure we track this down and correct it at that point.
Best wishes,
Vicky