Hi, everyone!
I have been trying to do a sequential assignment of the protein I am working with, but right now I am struggling with the “cross-peak <=> amminoacid” predicted/prediction assignment .
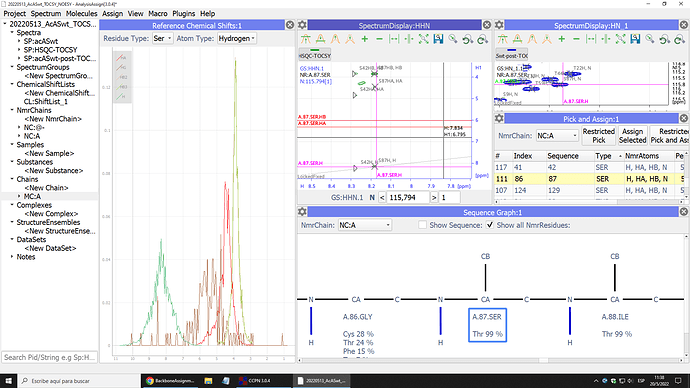
I am using an hsqc-tocsy, just to check if the coss-peak i believe is (i.e.) a Serine; really looks like one in my experiment. Fortunatelly, that is indeed what happens, yet for some reason that escapes me, the predicction algorithm resolves that those peaks belong to a Theonine or other AA. In the picture, you can see the Serine reference on the left, the tocsy assigned cross-peaks looking like a serine in the middle, an hsqc reference on the right, and the Sequence Graph at the bottom; predicting the threonine where a serine should be.
So, I am doing something wrong? I can not go any further with the sequencing (I have also an hsqc-noesy for that purpose), if I first can not assign the right AA to the right cross-peak.
Thank you in advance for any tip/aid/guide you can give me, I will really appreciate it.
Best regards!
Fran.
Hi Fran,
the sequence predictions are really only a guide, so it shouldn’t really matter what those are. In general, it is the Ca and Cb resonances that give you the most information about the AA type. So if you are not using 13C spectra, then it isn’t surprising if the predictions are not particularly accurate.
So I wouldn’t worry and would go with your own judgment based on the spectra and the chemical shift predictions.
As to the reason why the program is so convinced it is a Thr: I suspect this is because of the atom names you have used.
A Threonine has an alpha, a beta and three gamma methyl hydrogen atoms. These are referred to as HA, HB and HG% (the % in HG% indicating that all three methyl protons have the same chemical shift).
A Serine has an alpha and two beta hydrogen atoms. These are referred to as HA, HBx and HBy if the two beta chemical shifts are distinct and as HA and HB% if the two beta chemical shifts are degenerate (as in your case). If you can assign your two Serine beta hydrogens stereo-specifically they would be HB2 and HB3 - but it is relatively rare to make stereospecific assignments for these protons.
I suspect that if you rename your Serine HB to HB%, the prediction will change to Serine.
For a full list of all the IUPAC/NEF atom names see our NEF Atom Names page which should help you use the correct atom names and then get better predictions.
Best wishes,
Vicky
1 Like
Dear Vicky,
thank you so much for your fast reply! It really helped. I will try to do as you suggest, and correct the assignation  , and see what happens. In any case, thanks for your guidance.
, and see what happens. In any case, thanks for your guidance.
Kind regards,
Fran.
 , and see what happens. In any case, thanks for your guidance.
, and see what happens. In any case, thanks for your guidance.