Hi Everyone!
I`ve decided to take CCPNMR V3 for a whirl and assign a protein instead of with CARA (a transition from Windows to Mac). :)
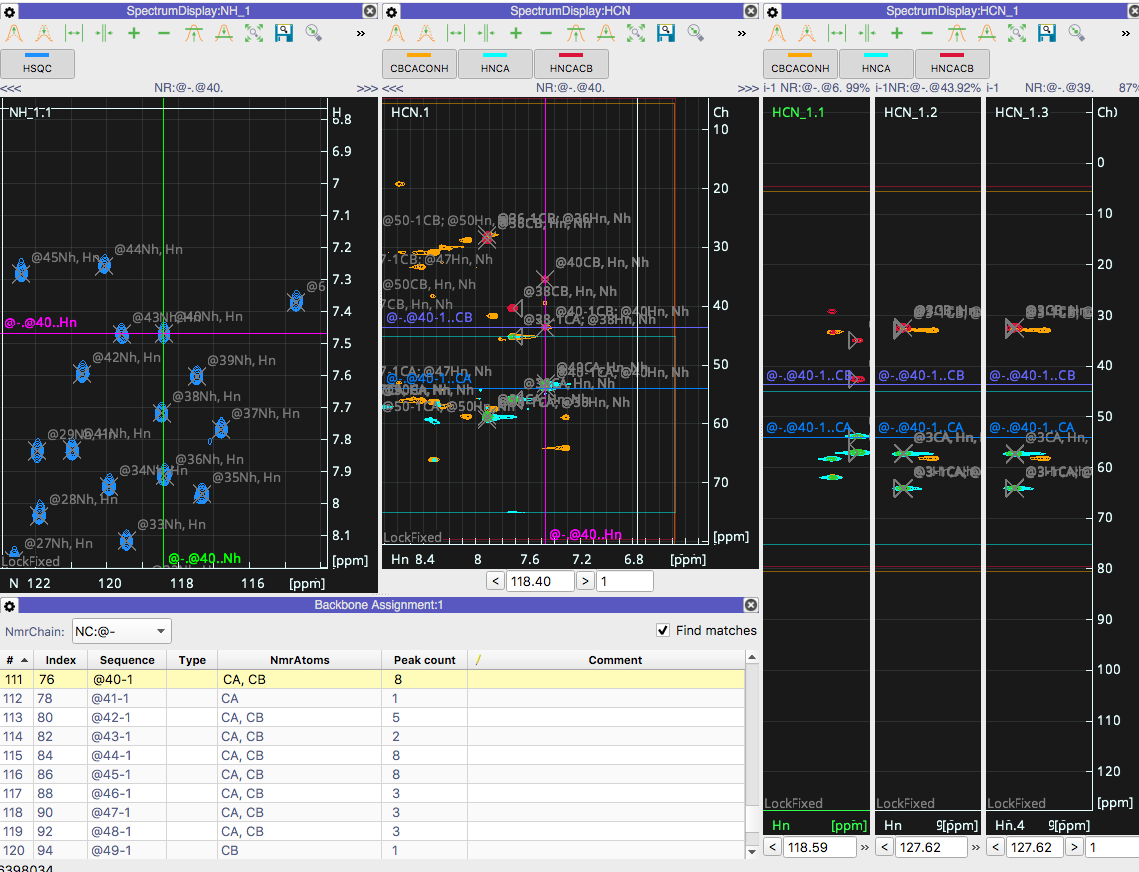
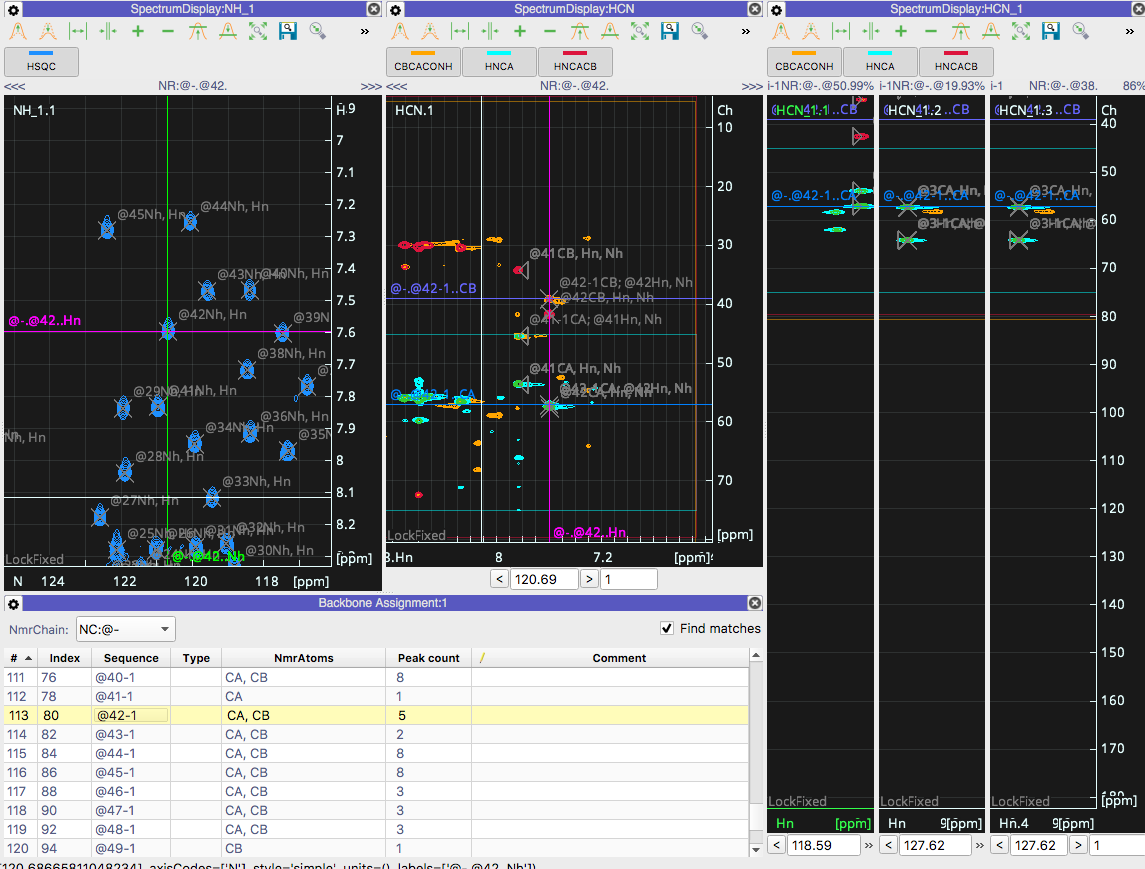
Following the Backbone assignment tutorial (using my own data) I can get as far to have all peaks picked in 3D and the Backbone assignment module is indeed matching strips but... Instead of moving each strip to the peak position in Proton AND Nitrogen it does not move in the Nitrogen dimension. See the attached screenshots which shows the matches `40-1` to `6` and `42-1` to `50` but there is no change to the Nitrogen position.
I would like to note that loading the tutorial data and following the guide works as expected with the backbone assignment matches moving to the correct nitrogen chemical shift. So I went ahead and checked the peak lists of my data and everything there appears to be correct. :/
Therefore, I assume this is just an issue of my settings, but I`m not sure where to look. This was a clean install of CCPNMR V3 beta 5 that has been updated twice (127 updates first then 5 updates the second time). Also, the gear icons on the Spectra view nor the Backbone Assignment modules appear to have anything pertaining to locking of a dimension.
I hope is just a simple tweak to my settings or maybe I`m doing something seriously wrong? I can always start again from scratch but was wondering if anyone could provide some advice?
Thanks,
Gage
Hi Gage,
I`ve not come across this before, even when working with other data. But I wonder whether it could be something to do with the Axis Codes (I note your HSQC Axis Codes seem to be H and N, the other ones are Hn, Ch and presumably Nh). You can check the Axis Codes for each spectrum by double clicking on the spectrum in the sidebar to bring up the Spectrum Properties dialog and then look in the Dimensions tab. Perhaps best to make sure that all your Axis Codes are the same across all your spectra. I generally leave them all as H, N and C, though I am aware that at the moment setting an experiment type will change your Axis Code (something I am keen to change...). In fact, the Backbone Assignment routine will work even if you don`t set experiment types.
Vicky
Hi Vicky,
That might be the issue but I`ve decided to complete assignments in CCPNMR V2 and after a trial run it seems like it is effortless. After completing the peak picking I will save the project folder and copy it over to open with V3. Maybe things will be set up properly and Backbone assignments will work as intended? I`ll keep everyone informed.
As for the axis Codes they are as follows:
HSQC - Nh, Hn
HNCO - CO, Hn, Nh
HNCA - Ca, Hn, Nh
HNCACB - C, Hn, Nh
CBCAcoNH - C, Hn, Nh
I will set these to all be C, N, and H and test the functionality as well.
Thanks,
Gage
Hi Gage,
Thank you for reporting the issue. We have been looking at this and seems to be independent from the Axis Codes. At the moment the `Navigate to Position` feature has an ambiguity when an NmrResidue `i` exists in different offsets (E.g. i-1, i+1 etc) so when you double click the i-1 it still goes to the `i` position; in your case, in the HSQC it goes to 42 instead of 41. We shall push an update as soon as we fix this issue.
Thanks,
Luca