Hello!
For the backbone assignment, the sequence graph module shows up with the schematic residue chain and the sequence. However, under the schematic, there are no predictions shown for the residues. Is there something in the settings that I need to change? I have checked the tutorial and have followed the directions to set up the different modules (for the most part). I had worked on a tutorial in the past and remembered seeing the residue predictions underneath the schematic.
Thanks for the help!
Domarin
Hi Domarin, we noticed that issue and we fixed few weeks ago. Have you tried to update recently?
Please run the update twice. Close and restart the program in the between. (One will update the updating mechanism!)
Regards
I updated again (twice) and still don`t have the predictions popping up. I am also having a problem with exporting peak tables. I am running the program through linux.
Hi Domarin,
I`m surprised you are still having trouble. I have just tried downloading the software again and applying the updates and both the prediction and table export seem to be fine. That is on a Mac, rather than Linux, but I would be surprised if that kind of thing would be platform dependent. Unfortunately, I don`t have access to a Linux machine at the moment to try that.
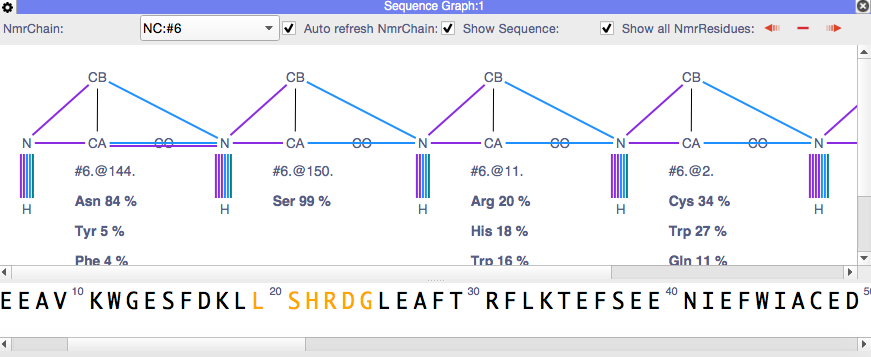
The AA type predictions you should be getting are both for each single NmrResidue and then also possible matches to the sequence once you have linked several NmrResidues (see figure). This works for me both for the Sec5 tutorial which you can download from the website and for other data. Which predictions are you missing? Could it be that you haven`t assigned your carbon NmrAtoms yet? Or do you need to change the NmrChain which is selected in the Sequence Graph module?
Vicky
Hi Vicky!
I opened up the tutorial and saw that the residue AA predictions pop up along with the peak assignments. When I open up my file, I am unable to see the peak assignments or the AA predictions. I have assigned Ca, Cb, CO, H, and N. Does it make a difference if the assignment is H or Hn? Subsequently for N or Nh? I have the correct things checked in the settings as well as changing the chain option.
Thanks!
Domarin
Hi Domarin,
it is indeed an issue with NmrAtom names. If the HN group atoms are Hn and Nh, then you don`t get predictions whereas you do, if they are H and N!
For the time being the best solution for you is probably to rename your NmrAtoms using the command line.
Type Space-Space in order to bring up the Python console. Then you`ll basically need to type in the following lines for each NmrResidue
get(`NA:@-.@1..Hn`).rename(value=`H`)
get(`NA:@-.@1..Nh`).rename(value=`N`)
Perhaps you can use copy/paste in a text file first to create all these lines and then copy/paste them into the console.
I had been thinking that it didn`t make sense for the NmrResidue Setup routine to name the HN atoms Hn and Nh, anyway (as those aren`t the NEF/IUPAC atoms names). So we`ll sort that out once everyone is back from their holidays. And for the time being it is probably best to do the NmrResidue Setup without setting the experiment type (in which case the atoms are names H and N!).
Vicky
Woops - sorry about those emoticons inserted automatically by the forum software! It`s
get(`NA: @ -.@1..Hn`).rename(value=`H`)
get(`NA: @ -.@1..Nh`).rename(value=`N`)
Without the spaces around the @ symbol!
V.
Hi All,
just FYI , the NmrResiduePopup creates nmrAtoms automatically with the same names as the spectrum axisCodes. You can change this on the spectrum popup (double click on a spectrum on sidebar). This obviously will not change now your nmrAtoms.
This is a quick macro to rename all your H and N nmrAtoms in the project.
Copy and paste this on the Python console and press enter:
=========
from ccpn.core.lib.ContextManagers import undoBlock
names = {`Hn`: `H`, `Nh`: `N`} #{old:new}
with undoBlock():[na.rename(names.get(na.name)) for na in project.nmrAtoms if na.name in names]
========
Close and reopen the SequenceGraph.
I hope this help for carrying on.
Best,
Luca
Hi Vicky and Luca!
I tried the command to rename all of the H and N nmrAtoms and I think it kinda worked. At least I see that some of the residues have predictions popping up beneath the name. I wet ahead and changed the spectrum axis codes in my experiments before running the macro. Interestingly, when I first was using the program and would input my spectra, I would change the spectrum axis codes but the software would not update it. I would open the spectrum window again and see that and HSQC would revert back to Hn and Nh. I don`t have that problem this time.
I am going to look through my assignments and see if something is missing as to why some of the predictions are not popping up.
Thank you both for all your help! It is much appreciated : )
Domarin