Hello,
I assigned peaks and then changed the assignment (from HB to HB% or from HB2/3 to HBx/y for example). However, somehow all the atoms remained. I checked each Peak Table and cannot find peaks with the old assignment. As a consequence ccpn is unable to predict residues.
Do you know how to solve it? I am using ccpn 3.1.1.
Thank you very much in advance.
Best,
Gabriella
Hi Gabriella,
first of all, you need to be aware of the difference between changing an assignment and changing an NmrAtom (i.e. the peak label). Changing the assignment means using a different NmrAtom/peak label for a particular peak and will only affect that peak. Changing an NmrAtom will affect your project globally and change that peak label in every place where it is used (i.e. also on other peaks and in the Chemical Shift List).
In the Peak Assigner (AP) you would change the peak assignment, by double-clicking on an NmrAtom to remove or add it to that peak’s assignments. Changing the NmrAtom would involve editing the NmrAtom, e.g. with Edit.
I’m unclear what you mean with “somehow all the atoms remained”. Which atoms remained where? Apparently in the PeakTables they did not remain? Perhaps you could explain in a bit more detail. It is unclear to me whether you were trying to change the assignment or the NmrAtoms and how the behaviour you got differed from what you were expecting.
I’m also surprised what you say about the program not being able to predict residues (amino acid types is presumably what you actually mean?). The use of HB vs. HB% etc. shouldn’t really affect this. However, we did make some changes to the amino acid type prediction a few months back, and it is possible that there could be bug in there. So it would be good to know how you changed the NmrAtoms names in an NmrResidue and how this then affected the predicted amino acid type.
Thanks,
Vicky
Hi Vicky,
Thanks for your fast answer! So sorry for the unclear explanation. I hope with the picture is a bit clearer :)
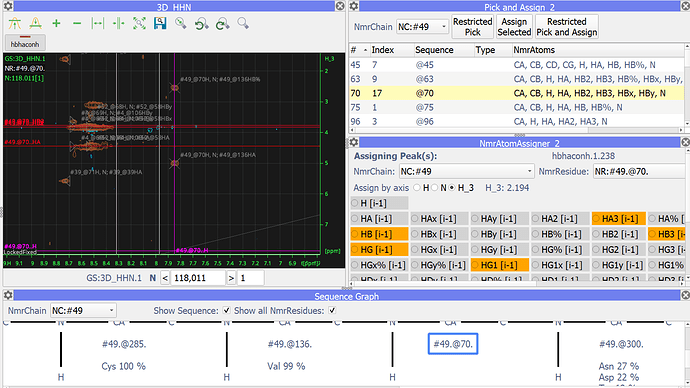
I was assigning peaks for the HBHACONH, I first used HA/HB or HA2/HA3 etc. then I changed them in HA%/HB% or HAx/HAy etc.
In the pick and Assign all the NmrAtoms remained, even though I changed the labels. So for example, in the upper right side of the picture @70 has both HB2,HB3 and HBx,HBy. Which I thought was causing then ccpn to not being able to predict the amino acid type (bottom of the pic).
Hi,
thanks for this.
It looks to me like there might be multiple issues going on here.
One is the behaviour of the NmrAtom Assigner. Basically, when you change the assignment, any previously created NmrAtoms remain part of the project, even if they are now no longer used.
Whether that is desirable behaviour is probably debatable: some people may want that, others may not. I’m tempted to say that it should tidy up and remove unused NmrAtoms. But we’ll have to discuss this as CCPN team (which may take a while, especially with people going on holiday over the next month or so).
The easiest way to remove these obsolete NmrAtoms is simply to go to Macro → Run CCPN Macros → removeUnusedNmrAtoms .
A separate issue is the prediction algorithm which does indeed seem to get a bit confused with side-chain H assignments. This might again take a weeks before it gets fully sort out, I’m afraid. We’ll keep you posted.
Vicky
Hi Vicky,
Thank you very much! Actually, once I removed the unused NmrAtoms the prediction of the residues worked again :)
Gabriella
1 Like