Dear all,
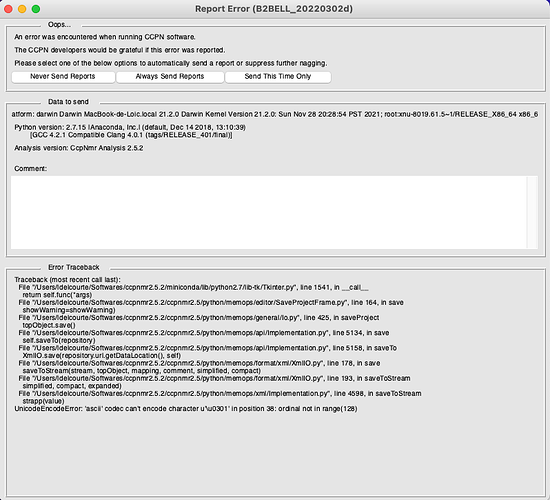
I am having some error while trying to save my project. Here is the message:
This only happens when saving after changing/adding atom assignment in one of my spectra (and only this peculiar one, a HCH-NOESY). I don’t think the problem comes from the spectrum, as I reloaded it and opened it in a new project, and there was no problem at all.
The error traceback would make me think it’s because of some encoding error (like for instance an “é” in the project name) but I can’t understand how it would be different before/after messing around with atom assignments.
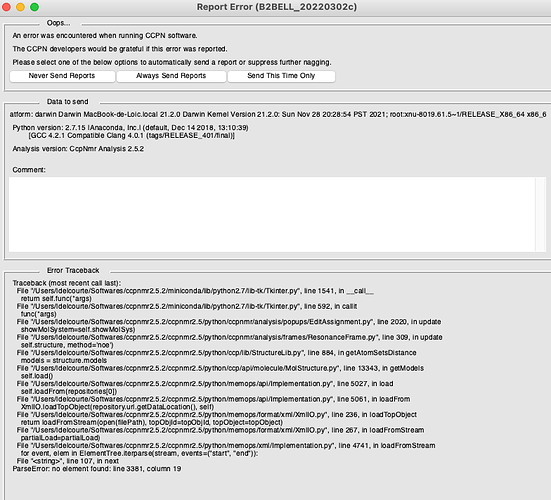
I also encountered a second error, occuring whenever I try creating a new peaklist for this spectrum: as soon as I add an peak and assign it (in the new peaklist), the software displays this:
If anyone has already seen this and knows how to solve it, it would help greatly.
Best regards,
Loïc Delcourte
Hi,
This is indeed a unicode error with the accent character.
https://www.fileformat.info/info/unicode/char/0301/index.htm
I guess somehow a character with an accent has got into the project.
The fix is clunky - analysis project files are stored in plain text XML format so you can use a text editor to find and remove the offending character. Do make a safe archive (e.g. tar.gz on linux/mac .zip on windows) of your project before you attempt this in case you break it.
Do let us know where you find the rogue character of get in touch if you can’t solve the problem.
Brian
PS the second error looks as if one of the project files may be truncated. see https://www.jiscmail.ac.uk/cgi-bin/wa-jisc.exe?A2=CCPNMR;6f3c3a9c.2108. You may need to go back to an earlier version of your project.
Hi Brian,
Thanks for your help. Unfortunately I didn’t find any non-unicode character in the xml file (which kinda makes sense in my opinion, since I can still save the project as long as I don’t change anything in this particular spectrum).
As for the truncated problem, the xml ends with the same lines as all of my previous project saves. Some of the earlier saves also have this problem (from like 2 weeks ago).
Thanks for your help,
Loïc
Interesting update: I think the whole thing may actually be linked to the pdb structure I added. When I try to open the structure menu/window, I actually get the same error (second screenshot of the original post).
Is there a way to delete the structure in any of the files from the project folder ? In which one should i look for the structure ?
I tried removing the MolStructure and Validation folders in Project/ccp/nmr/molecule but it didn’t change much.
Update 2: it seems like everything is working now.
I couldn’t pinpoint what was causing this issue, but deleting the structure from a previous (but still very recent) save, re-importing the spectrum and then re-importing the structure seemed to work well.
My only explanation would be that there was some conflict between the structure and the predicted peaklist I made for the spectrum.