Hi there
Thank you for getting back to me regarding my previous post. I don`t think I was using the linker bonds in the correct manner.
However I am wondering how the chembuild software fits together with the CCPN analysis software. I can export modified residues to XML files but how do I actually import these into analysis? Where am I supposed to export these XML files to so that they can be used in a CCPN project?
Additionally, exporting any structure as a CCPN XML file does not work with this message showing up in the python terminal:
Traceback (most recent call last):
File`/Applications/CcpNmrLauncher.app/Contents/Resources/ccpnmr3.0.0/src/python/chemBuild/ccpnmr/chemBuild/gui/ChemBuildMain.py`, line 1131, in exportChemComp
stream = open(streamPath, `w`)
FileNotFoundError: [Errno 2] No such file or directory: `/Users/thomas/Files/SGIGS3.ccpn/Ser1/protein+Ser1+ChemBuild_thomas_2020-02-07-15-04-11-346_00001.xml`
Exporting to any other format seems to work fine but I have found a workaround for xml files: if you create a folder with the name you want to give your exported structure (file name not the name under compound info) and place it in the same directory you`re trying to export to a CCPN xml file is successfully created.
(Assuming this is a bug and I`m not just using the software incorrectly again!)
Many thanks
Thomas
Hi Thomas,
thanks for this. I`ll take a look at it, but may not be able to get back to you till Monday.
In the mean time, it is worth pointing out that to do assignments, you don`t actually need to import a molecule into the programme as you did in V2.
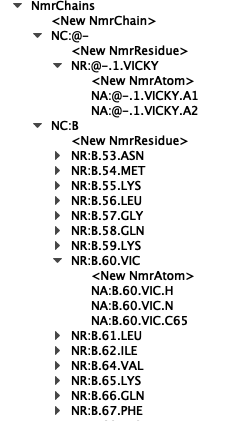
When you make an assignment you can just give your NmrAtom/NmrResidues any name you like (see attached Figure - double click on NmrAtoms/Residues/Chains to edit them). I think the only time you will run into problems giving them bespoke names is when you have created an NmrChain (with NmrResidues and NmrAtoms) from a Chain (and to my mind that is probably a bug - we`ll have to look into that).
So if you wanted, you could create your bespoke molecule in advance and then you will have the NmrAtoms already there to assign to your peaks, or you can just create the NmrAtoms using your nomenclature as you make your assignments (e.g. in the Peak Assigner).
This is one of the ways in which V3 is hopefully a bit easier to use and less rigid in its working than V2.
Vicky
Hi Thomas,
finally able to get back to you on this. This is going to be quite long, so I hope you will bear with me.
As yet, ChemBuild and Analysis V3 are not properly integrated. And I suspect it will take us a little while to get this sorted properly. There are one or two workarounds you can use, but it really depends what your ultimate aim is (why are you trying to import what).
Analysis V3 is currently able to read pdb files and SMILES, but not the CCPN ChemComp .xml files and not mol2 files.
pdb files can simply be dragged in, and will then go into the `Structural Ensemble` part of the data structure (note that is only recognises/read/does something with ATOM and not HETATM lines). Once they are in, there is not an awful lot you can do with them at the moment, though. It would make sense to be able to generate an NmrChain from a Structural Model, and I shall add that to our ToDo list.
SMILES notations (not files) can be entered in the Substances section:
- double-click on <New Substance> under Substances in the sidebar
- Click on `Show more`
- copy/paste your SMILES into the Smiles box
- Click Apply or OK and the molecule will be added
ChemBuild will write pdb files, but not SMILES. If you want your ChemBuild molecule in SMILES notation, you will have to export it as mol2 and then use OpenBabel (http://openbabel.org/wiki/Main_Page) to convert it to SMILES.
Thanks for pointing out the issue with writing xml files. We shall certainly try to fix that. Though interestingly I note that the manual says `At present ChemBuild will not export ChemCompCoord XML files; the atom coordinate records used by CCPN templates. This will only be introduced after better 3D molecular dynamics.` ChemBuild was originally written by Tim Stevens who left CCPN some time ago and we haven`t done much to it since. Though we are intending to use it more in future.
Finally, on to thinking about why you might be wanting to import something from ChemBuild into V3 and the implications for what you might want (and be able) to do:
a) If you are wanting to import a small molecule (i.e. not a protein, peptide or DNA/RNA), then I would suggest importing as a SMILES. You can then generate an NmrChain from your substance (double-click on <New NmrChain> and select Clone from Substance). You can then assign your molecule using the NmrAtoms in that NmrChain (the molecule will go into a single NmrResidue in the NmrChain).
b) If you have a non-standard amino acid in your peptide/protein, the easiest thing is probably to enter the chain using the natural amino acid most closely related to your non-natural one. Create an NmrChain from your Chain (double-click on <New NmrChain> and select Clone from Chain). Then delete/add NmrAtoms to the non-natural amino acid to create your non-natural amino acid. You can then use the NmrAtoms to do your assignment. The only drawback with this is that the program will not be able to recognise your non-natural amino acid as being assigned, because there is no equivalent in a Chain. At the moment it isn`t possible to modify Chains (except delete residues), but we will be adding such features in future.
What is worth noting is that you can make quite a few changes to your NmrChains and NmrResidues (by double-clicking on them in the sidebar). So you can import a SMILES as a substance, create an NmrChain from this (the whole molecule will go into a single NmrResidue) and then double click on the NmrResidue and change the NmrChain, Sequence Code (i.e. Residue Number) and Amino Acid name to those of your choice, and this will be slotted into another NmrChain. (So you could have another NmrChain in which you had removed an NmrResidue and then you could replace it with something from a SMILES in this way.)
Sorry this is so long and that these things are not yet fully developed in V3, but I hope this gives you some clues as to how you can proceed, depending on what you are trying to do.
Best wishes,
Vicky