Hi Eliza @ElizaP
I am having some difficulties with compatibility between v3 and getting CYANA to read and write .nef files. As far as I can tell, CYANA can read .nef files but gets stuck on recognising the dimensions of 1H-1H NOESY spectra (copied below). Do you have any tips on how I need to edit my .nef files to get CYANA to read them?
Thanks,
Anne
cyana> read nef CX0001_CCPproj1.nef
*** WARNING: Unsupported NEF format version `1.0`.
*** ERROR: Cannot read line 277 in frame `nef_nmr_spectrum_CX0001_NOESY_pH6_298K`:
1 ppm 1H 700.4732885 9.99724 9.775281268 circular true . H1
NMR exchange format file `CX0001_CCPproj1.nef` read.
Hi Anne,
Cyana (cyana-3.98.13 version) needed few changes in NEF file to read it. All spectra information that are written in NEF file need to agree with cyana-3.98.13/lib/cyana.lib file.
In your exported NEF file:
1. In the saveframe for your spectra (for instance: save_nef_nmr_spectrum_2dnoe`1`) you should change _nef_nmr_spectrum.experiment_type tag to use Cyana nomenclature from cyana.lib file.
For 2d NOESY it should be:
_nef_nmr_spectrum.experiment_type NOESY
2. In the `_nef_spectrum_dimension` _loop _nef_spectrum_dimension.axis_code also have to be changed (again in accordance with cyana.lib).
It should look like this:
loop_
_nef_spectrum_dimension.dimension_id
_nef_spectrum_dimension.axis_unit
_nef_spectrum_dimension.axis_code
_nef_spectrum_dimension.spectrometer_frequency
_nef_spectrum_dimension.spectral_width
_nef_spectrum_dimension.value_first_point
_nef_spectrum_dimension.folding
_nef_spectrum_dimension.absolute_peak_positions
_nef_spectrum_dimension.is_acquisition
1 ppm H1 749.7835083 16.67147898 13.12757051 circular true .
2 ppm H2 749.7835083 13.33718319 11.46041975 circular true .
stop_
3. To avoid NEF version warning you should change:
_nef_nmr_meta_data.format_version 0.91
This tag is the first saveframe (save_nef_nmr_meta_data)
Hopefully this should help.
If you have any questions please let me know.
Eliza
(02-03-2021, 12:24 pm)ElizaP Wrote: Hi Anne,Thanks Eliza!
Cyana (cyana-3.98.13 version) needed few changes in NEF file to read it. All spectra information that are written in NEF file need to agree with cyana-3.98.13/lib/cyana.lib file.
In your exported NEF file:
1. In the saveframe for your spectra (for instance: save_nef_nmr_spectrum_2dnoe`1`) you should change _nef_nmr_spectrum.experiment_type tag to use Cyana nomenclature from cyana.lib file.
For 2d NOESY it should be:
_nef_nmr_spectrum.experiment_type NOESY
2. In the `_nef_spectrum_dimension` _loop _nef_spectrum_dimension.axis_code also have to be changed (again in accordance with cyana.lib).
It should look like this:
loop_
_nef_spectrum_dimension.dimension_id
_nef_spectrum_dimension.axis_unit
_nef_spectrum_dimension.axis_code
_nef_spectrum_dimension.spectrometer_frequency
_nef_spectrum_dimension.spectral_width
_nef_spectrum_dimension.value_first_point
_nef_spectrum_dimension.folding
_nef_spectrum_dimension.absolute_peak_positions
_nef_spectrum_dimension.is_acquisition
1 ppm H1 749.7835083 16.67147898 13.12757051 circular true .
2 ppm H2 749.7835083 13.33718319 11.46041975 circular true .
stop_
3. To avoid NEF version warning you should change:
_nef_nmr_meta_data.format_version 0.91
This tag is the first saveframe (save_nef_nmr_meta_data)
Hopefully this should help.
If you have any questions please let me know.
Eliza
This solved my problem and I can now write .prot and .peaks files from my .nef file.
Hello Anne and Eliza,
Thanks for the above information - I had similar problems importing NEF file into CYANA.
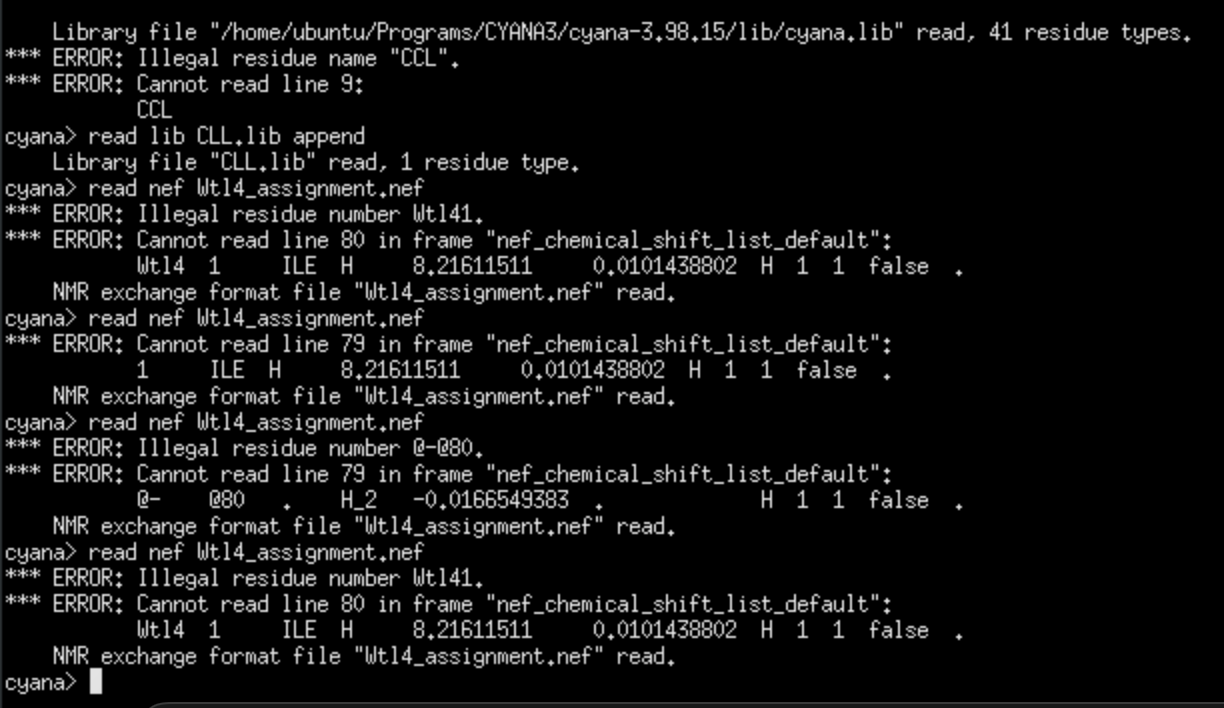
I have my 1D proton, TOCSY and ROESY spectra imported into CCPN to pick the peaks in mainly TOCSY and ROESY spectra. I cleaned up the chemical shift list before exporting to remove the same shift picked in different spectra, and I left some of the aromatic peaks unassigned. I only exported the chain, the chemical shift list and the ROESY peak list. When I was importing the NEF file, CYANA could not read the chemical shift list (see the screenshot).
Thanks for any help in advance!
Hi,
Cyana will not read anything with “@” sign, so I usually make a duplicate of the chemical shift list and remove everything that is not standard, but that also means you may need to change which chemical shift list is used for your spectra.
If you can send your NEF file (support@ccpn.ac.uk) I am happy to try it myself.
I have never tried to read 1D peak list from NEF in Cyana, and looking at Cyana library I am not sure if it has provision for 1D spectra, but perhaps it does.
Kind regards,
Eliza
Dear Eliza, @ElizaP
I am fixing my nef file and right now I am stuck with this error:
I have tried the following changes:
_nef_nmr_spectrum.sf_framecode nef_nmr_spectrum.15Nnoesy-52 NOESY
_nef_nmr_spectrum.sf_framecode _nef_nmr_spectrum.15Nnoesy-52 NOESY
_nef_nmr_spectrum.sf_framecode nef_nmr_spectrum.15Nnoesy-52
So far none of these worked. Could you please help me with this?
Thanks!
Carolina
Hi Carolina,
I tried the name you send and had no issues with the name of the spectra.
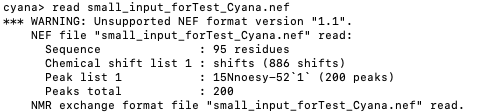
Those kind of errors usually occur when there are inconsistencies in the name of the saveframe.
If you can send your NEF file to us (support@ccpn.ac.uk) I am happy to have a look.
Best wishes,
Eliza
Hi Eliza,
Thank you very much for your quick response.
I had changed the name of the save frame to _nef_nmr_spectrum.15Nnoesy NOESY as you suggested to the initial post.
Now, I changed it back to the original name save_nef_nmr_spectrum_15Nnoesy1 and I get this error:
Thanks,
Carolina
Hi Carolina,
You can just hash (#) this line, then it should read.
Eliza