I’m trying to upload a minorly edited BRMB file that contains all the same information as the file you can download for BMRB, but the chemical shift list is changed. I’ve just taken the original BMRB file (that loads into CCPN fine) and edited the chemical shift list only. When I try to import the new list, all the information except for the CL is imported. How can I fix this?
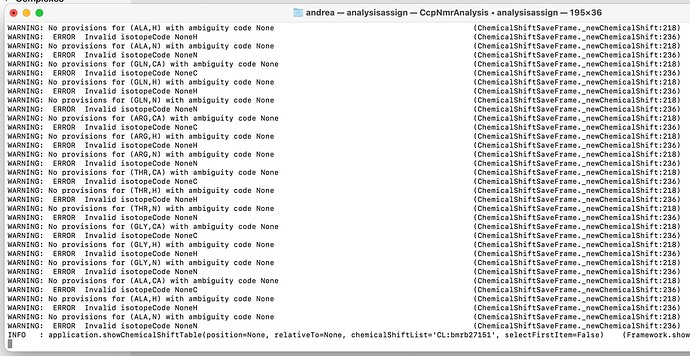
I attached an image of the error that I’m seeing that I don’t know how to fix.
P
Hi,
it depends how you changed the chemical shift list…
It looks as though you didn’t specify the isotope types correctly (H, C and N). See Assigned Chemical Shift File Help for an example of what the shift list should look like and where the isotope column is.
It seems you also haven’t specified any ambiguity codes - though that shouldn’t stop the import, I think. Our code seems to be expecting the ambiguity code to be 1, 2 or 3. You can find information about ambiguity codes at Assigned Chemical Shift File Help . For the H, N and CA atoms for which the errors are being thrown the ambiguity code would generally be 1.
Vicky
Thank you for the help! I fixed the formatting of my file. Now the error that Im getting is “Existing NmrResidue:test1.None.GLY does not match residue type ‘ARG’ (ContextManagers.catchExceptions:218)”’
I found and older thread Error with getting simulated HSQC from imported NMRSTAR file, and my issue is probably the internal residue clash that is discussed. GLY and ARG are the 1st and 3rd amino acids, but are 1 and 2 on the list because I don’t have a chemical shift value for residue 2 . The GLY is the only chemical shift that gets imported into CCPN, probably since it’s not connected to the 2nd amino acid.
Would I still need to upload each chain individually, or should 3.1.1 be able to upload all of them at once? Let me know if there is any other information you’d need!
Hi,
I don’t think there should be a problem with importing a chemical shifts with some amino acids missing. But it could be that there is mismatch between your sequence and the shifts you trying to import.
The easiest thing might be for you to send us the file you are trying to import and possibly also the project you are trying to import it into (if that already has something in it). Simply send them to support@ccpn.ac.uk and we can take a look and hopefully get to the bottom of this.
Vicky
I ended up switching the file format from the NMRstar to the NEF format and was able to upload the data with the entire chemical shift list without issue. Thank you for your help!
1 Like