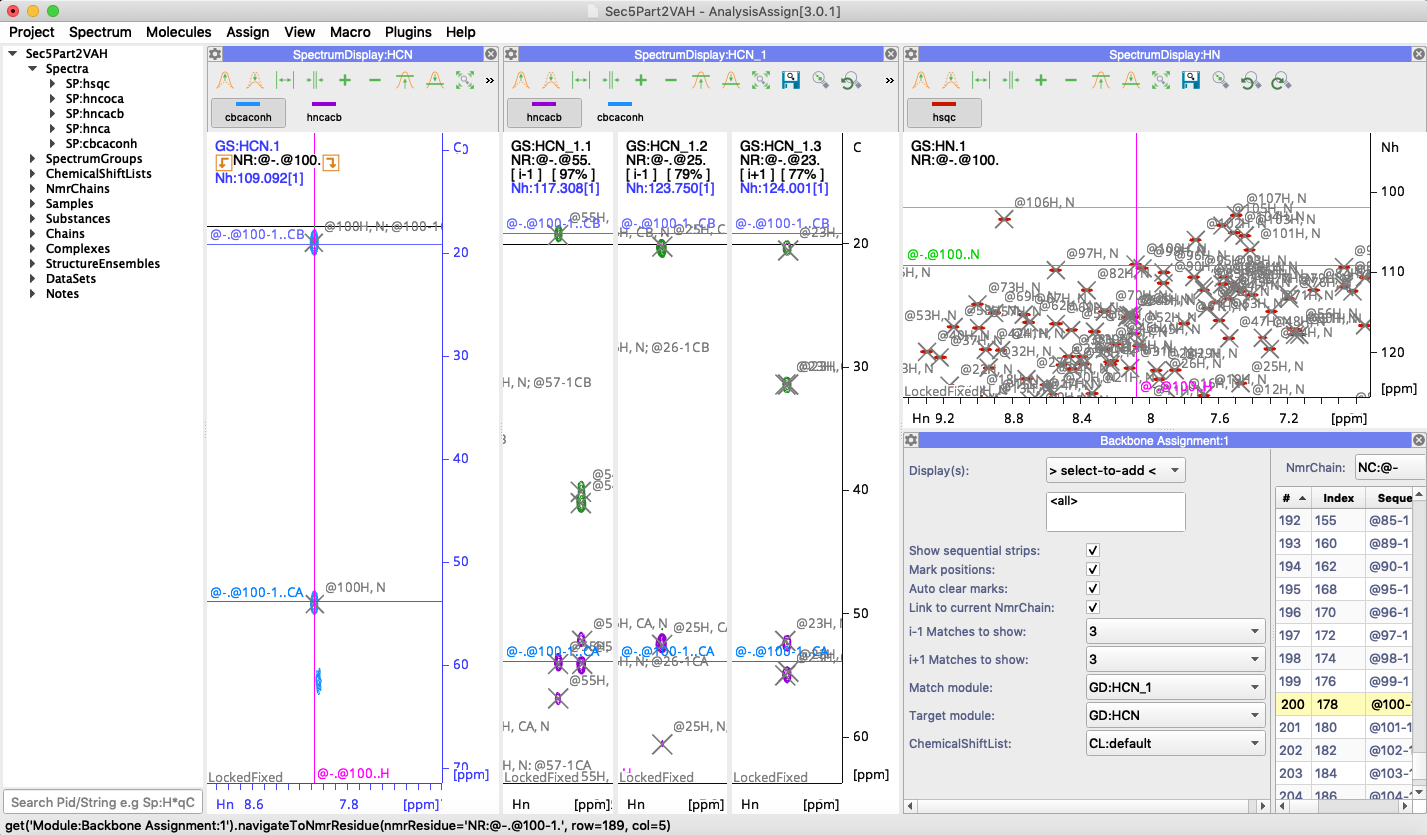
I was recently operating ccpnmr v3 on a beta version of NMR box v7 and it was working well. However, they just completed the update and I am now using my personal NMRbox page instead of the beta version and I am having issues with ccpnmr. In this version under backbone assignment it asks you to choose a match module and a target module, as opposed to just a match module. This new setting says that I am required to assign a spectra to it but it appears to be messing everything up. I no longer see the i-1 or the arrows at the top of the spectra to connect residues. It was previously working perfectly. What should I do? Thanks.
Hello,
I am using Ccpnmr V3.0.1 in MacOSX Mojave, besides the design is better in this version comparing to the version 3.0.0 (congrats for that!) the module backbone assignment is more confused. I suggest the use of two match modules (HNCACB and HNCO window`s) for one target module (HSQC window) and just the one of two possibilities: i+1 or i-1 and not both.
Thanks,
Micael Silva
Hi,
sorry not to get back to you about this sooner. See attached for the way in which I usually set this up. The Target module also needs to be a 3D - the one in which you want to see your little arrows for connecting. I`ve actually thought it might be better to rename it `Search Module` - though perhaps someone else has a better idea.
Vicky
Hi,
Thanks for the suggestion. Sounds good when the target module is the match module duplicate. Anyway, should not be possible to have two match module`s at the same time? Sometimes the HNCO/ HNCACO helps the assignment.
Cheers,
Micael
Hi Micael,
yes, we`re working towards that. We have plans to add horizontal separators/tiling which will mean that you can have the HNCO/HNCACO open in the same modules as the CA/CB spectra and the strips will then be focussed on the CA/CB/CO areas separately and cut out all the space in between.
Vicky
Hi Vicky,
As it is, I used the backbone assignment two times (once for match HNCA/HNCOCACB and the 2nd for confirmation with match HNCO/HNCACO). Also, in the assignment inspector, the i-1 not become automatically the residue before (as example: V39H,N; A_39-1C become V39H,N; G38C just with NmrAtomAssigner review), this is really useful to obtain a Standard Deviation of that Chemical Shift.
Thanks and congrats for the amazing job with this software/ pipeline.
Micael
I also have problems during the backbone assignment with dropping on the curved arrows that replaced the arrows at the top of the windows. Every time I do this it seems to be an invalid.
Heiner
Hello Heiner,
make sure you are picking the strip up by the NmrResidue (e.g. NR:@-.@19.), not the strip ID (e.g. GS:HCN_1.1).
If that still doesn`t help, then please let me know what operating system you are using and also make sure you are on version 3.0.1 with all the updates installed (which will in fact take you onto V3.0.1.1).
Thanks,
Vicky