Hi Anna,
it should be possible to work with RNA in the program, although this isn’t something we have actively supported and tried much. Our main problem (apart from finding the time for it) is that we don’t have any RNA spectra. So if you or someone from your group were willing to share spectra with us and have a conversation about some of your needs, then we can start work on this more quickly and easily.
Now for what there is:
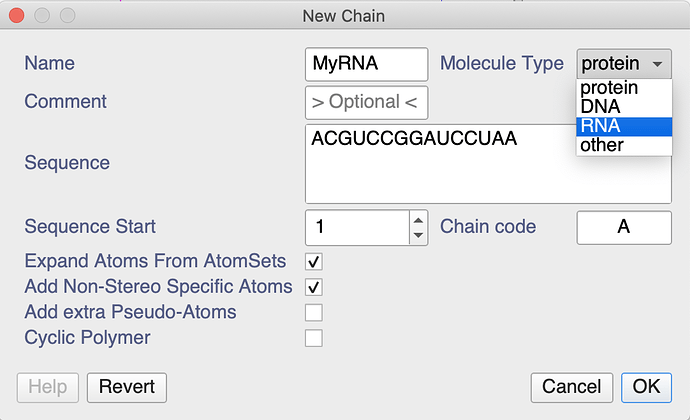
When creating a new chain, you can create both DNA and RNA chains. After double-clicking on in the sidebar you could do the following:
I notice that the drop-down menus, e.g. in the Peak Assigner don’t list the RNA atoms. That’s something we’ll have a look at, though it will have to wait until after our conference and 3.2 release next week. However, you can type whatever you want into the NmrAtom Name box and don’t have to use the drop-down menu. So just type whatever your atom is called.
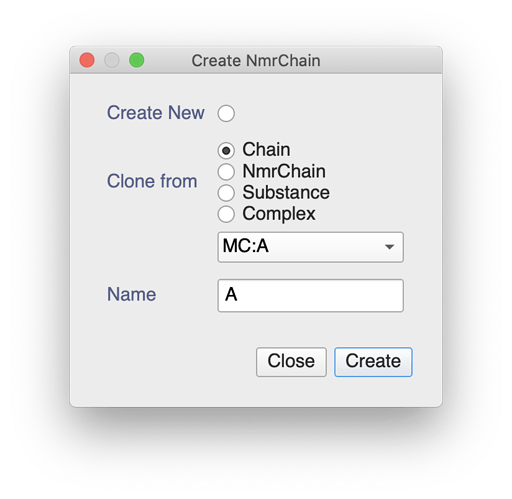
You can always create an NmrChain from your Chain, if you wanted. After double-clicking on in the sidebar you could do the following:
This will give you an NmrChain with all the NmrAtoms in your RNA molecule. You can use these to assign peaks by dragging from the sidebar on to a selected peak.
With regard to importing RNA chemcial shift lists and dihedral restraint lists: it depends a bit on what format your data is in. You can import BMRB and NEF files, but presumably your data are in a different format at the moment?
There are probably several routes you could take at this point:
- change your files into NEF files by hand. Not entirely recommended, as it can be error prone, but we certainly do this from time to time. We have a tutorial (How to create a NEF file form tabular data) which will give you some advice on how to do this at Tutorials - CCPN - Collaborative Computing Project for NMR .
- You could try to import your data into Version 2 using the Format Converter. If you then save your project, you should be able to open it Version 3 and then create the NEF file.
- @varioustoxins has written some software called NEFPipelines which can do some data conversion. We have a tutorial on that, but I don’t know what formats it can currently handle and what it does with RNA data. But I’ll ask him to take a look at this thread and let us know if it might be of any help.
The key thing, however, will be to know what format you currently have your data in.
But do remember: the software is completely flexible about what your NmrAtoms and NmrResidues are called, so although much of the software currently assumes you are working with proteins, you can name things as you like and most of the basic functionality will be fine.
Vicky