Hi,
I’m having a problem with my protein sequence assignment. When I go to M:Assignment:Protein sequence assignment:Spin system table, only a small portion of my total spin systems are visible. Even the ones that are in the table do not align very well between query and match windows.
I didn’t notice having any problems with referencing/assigning spin systems before attempting protein sequence assignment. Does anyone know what might be the problem?
Thanks,
Sarah
Hi Sarah,
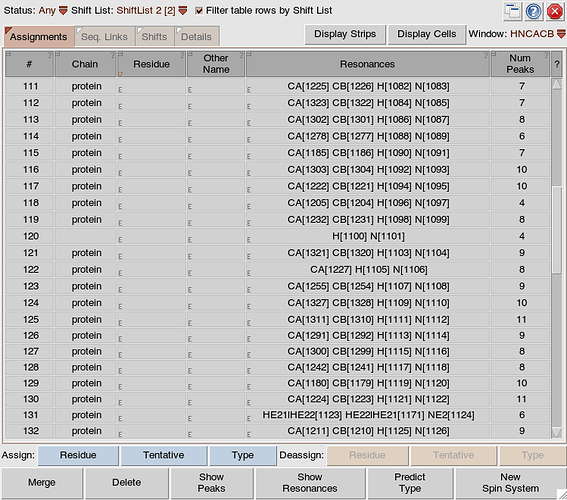
it’s hard to diagnose this at a distance, but I wonder whether it is to do with whether your resonances are all correctly specified. The best way to check this would be to go to M:Resonance:Spin Systems. I would expect it to look something like this:
i.e. for most spin systems you should have Resonance types H, N, CA and (depending on what type of spectra you are using) perhaps also CB and C.
Other things to check would be your Experiment Types. You could also try increasing the Tolerances a bit in the Windows&Spectra tab and see if that has an effect.
I suspect you are getting a low degree of alignment because the program is only working with a subset of spin systems, meaning the the correct match isn’t available, thus only low quality matches are possible and the alignment looks bad.
If you like, send us your project to support@ccpn.ac.uk and one of us will have a look and see if we can work out what is going on.
Vicky