Hello,
I use CCP 3.1.1, I can download and work with 2D and 3D nmr spectra without any problem. However when I try to load a 1D spectrum in the same way as I load a 2d or 3d spectrum (by selecting the 1r file in the experiment folder created by topspin), I can’t see any 1D spectrum but a straight line, with time in vertical axis and H in horizontal axis. I figured out that when I extract a 1D spectrum from a 2D spectrum with topspin and then upload it into ccp, i obtain my 1D spectrum, but when I try directly with a 1d spectrum, I can’t obtain any spectrum with ccp…
Do you have any idea of what I could do or how I could transform my 1D spectrum to obtain a spectrum with ccp ?
Thank you very much,
Béatrice
Hi Béatrice,
a couple of things:
-
would be great if you were able to upgrade to version 3.2.2 at Software Downloads - CCPN (I’ve just been in touch with someone whose project wouldn’t open in Version 3.1.1, but was fine in 3.2.2 - there are many bug fixes and other improvements in the new version).
-
It is definitely possible to open Bruker 1Ds in Analysis - most of our AnalysisScreen customers do this all the time (and have done for many years - so I doubt the version makes any difference here). I wonder if this is something to do with the processing in Topspin which somehow doesn’t mark your 1D intensity axis correctly - and AnalysisAssign then interprets it as a time axis instead. How we fix this and work out what you need to do differently I’m not quite sure, though. Can see you any difference between the extracted and the “native” 1Ds in Topspin?
Vicky
“how we fix this and work out what you need to do differently I’m not quite sure…”
One answer would be can you send us an example project and spectrum that has this problem and we can do some debugging?
regards
Gary
Thank you for your answers,
I unfortunatly couldn’t install the latest version of ccpn, it came with errors when trying to launch ccp with “assign” command.
I tried to compare the 2 1d in topspin but I did’nt find anything relevent that could help me to open it with ccp.
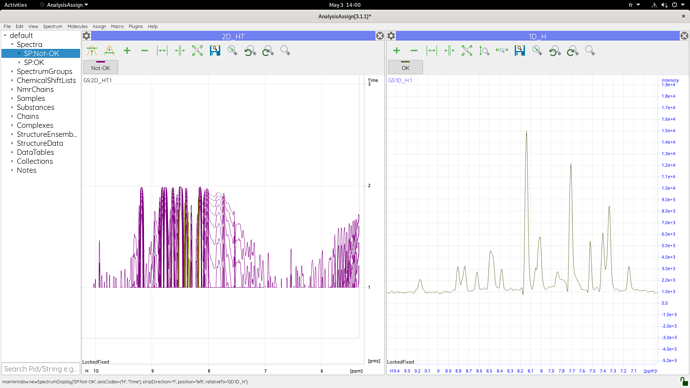
Here is a filesender link with 2 folders, one containing the 1D experiment that I can open properly with ccp, and the one I faced an issue with, and a screenshot of the 2 spectra in ccp.
https://filesender.renater.fr/?s=download&token=7682ed8f-2f82-4370-b609-de8b035f07e7
Regards,
Béatrice
hi Beatrice we will have a look at the files, could you send us the text of the errors you get when you try to run assign and information about what OS you are running and we can have a look at that as well…
regards
Gary
nb please note Monday is bank holiday in the uk!
Hi Gary,
Here is the full text I obtained, with the error at the last line.
root@GRE052304:/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/bin# ./assign
Program licence (non-profit) valid until Mon Jan 4 16:34:08 2100
Traceback (most recent call last):
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/runpy.py”, line 196, in _run_module_as_main
return _run_code(code, main_globals, None,
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/runpy.py”, line 86, in _run_code
exec(code, run_globals)
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/AnalysisAssign/main.py”, line 32, in
from ccpn.AnalysisAssign.AnalysisAssign import Assign as Application
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/AnalysisAssign/AnalysisAssign.py”, line 30, in
from ccpn.framework.Framework import Framework
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/framework/Framework.py”, line 71, in
from ccpn.core.IntegralList import IntegralList
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/core/init.py”, line 536, in
from ccpn.ui.gui.MainWindow import MainWindow
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/ui/gui/MainWindow.py”, line 50, in
from ccpn.ui.gui.lib import GuiStrip
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/ui/gui/lib/GuiStrip.py”, line 52, in
from ccpn.ui.gui.widgets.Frame import Frame
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/ui/gui/widgets/Frame.py”, line 74, in
from ccpn.ui.gui.widgets.ScrollArea import ScrollArea
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/ui/gui/widgets/ScrollArea.py”, line 33, in
from ccpn.ui.gui.lib.OpenGL.CcpnOpenGLDefs import BOTTOMAXIS
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/src/python/ccpn/ui/gui/lib/OpenGL/init.py”, line 53, in
from OpenGL import GL
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/site-packages/OpenGL/GL/init.py”, line 4, in
from OpenGL.GL.VERSION.GL_1_1 import *
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/site-packages/OpenGL/GL/VERSION/GL_1_1.py”, line 14, in
from OpenGL.raw.GL.VERSION.GL_1_1 import *
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/site-packages/OpenGL/raw/GL/VERSION/GL_1_1.py”, line 7, in
from OpenGL.raw.GL import _errors
File “/home/bvibert/Downloads/release3.2.2Ubuntu22.04-Academic/ccpnmr/miniconda/lib/python3.10/site-packages/OpenGL/raw/GL/_errors.py”, line 4, in
_error_checker = _ErrorChecker( _p, _p.GL.glGetError )
AttributeError: ‘NoneType’ object has no attribute ‘glGetError’
My OS is a Debian GNU/Linux 11, 64-bit, so I tried to install the version for Ubuntu 22 (maybe it wasn’t the best).
Thank you for helping!
Béatrice
hi beatrice
thank you so much for the information. could you also type this in a terminal and send us the text that comes out it, will allow us to see exactly which version of ubuntu you have. (note we may need to come back and ask about hardware and drivers at a later date if this doesn’t help)
lsb_release -a
for an example of what you should see go to
How to check your Ubuntu version: a guide - IONOS.
regards
Gary
Hi again Gary,
Here is the text that comes out :
No LSB modules are available.
Distributor ID: Debian
Description: Debian GNU/Linux 11 (bullseye)
Release: 11
Codename: bullseye
I hope it could help, don’t hesitate if you need any other information,
Regards,
Béatrice
Hi Beatrice,
I’ve found the problem with your 1D. For some reason your pdata/1 folder included the following two files: proc2 and proc2s
As soon as you remove these two files the 1D opens as normal.
I’m not quite sure why those files were there (they seemed to include lines with Mdd… - had you been processing the data with MDD?). Normally they should only be there for a 2D data. For that reason AnalysisAssign assumed that the data was 2D data and then got confused and did strange things.
Vicky
Hi Vicky,
Great ! Indeed, when I remove these two files it works perfectly.
Thank you very much for your help,
Béatrice
Hi Béatrice,
Do you know how those additional files might have ended up there? Had you copied a directory, or done some other processing? We’re just wondering how likely anyone else is going to end up with this situation and whether we need to change our algorithm which loads Bruker data (or at least improve some of the warning messages). Detecting the dimensionality of a Bruker spectrum is (bizarrely) not straight forward: there is no parameter which specifies this anywhere!
Thanks,
Vicky
Hi again Vicky,
I’ve tried with different experiment types to try to understand how theses files appear, but I can’t find any logic. I always have these 2 files, even if I just create a new experiment using a bruker 1D parameter set.
Thanks,
Béatrice
Okay - thanks, Béatrice.
In that case I’ll see if we can change the logic for how we work out and test what dimensionality a Bruker spectrum is.
Vicky