Hi there,
I seem to have made a little bit of an oopsie. There appears to be a deletion in my chain A at residue 219 - this essentially shifts everything after this deletion up one residue. I know editing a chain is not possible so I created a chain B.
But I am unsure how to transfer the assignments over with the sequence renumbering correction in the middle of the chain. Is there a way to maybe manually do this in the NEF file?
Thanks!
Hi,
In order to give you the most applicable solution could you clarify whether you are fixing your Chain (created from the imported sequence) or your NmrChain (created as you made your assignments)?
Have a look at https://forum.ccpn.ac.uk/t/chain-starting-number-once-the-chain-is-already-created/1277 there are methods for both the Chain and NmrChain objects that should allow you to fix the residue numbering in a few lines in the python console (space, space to open) or as a macro. Then it sounds as if once fixed, you could then delete your existing (gappy) Chain A and change your correct Chain B to be called Chain A and all should be well.
Make sure you have saved and archived first though!
Good luck
Brian
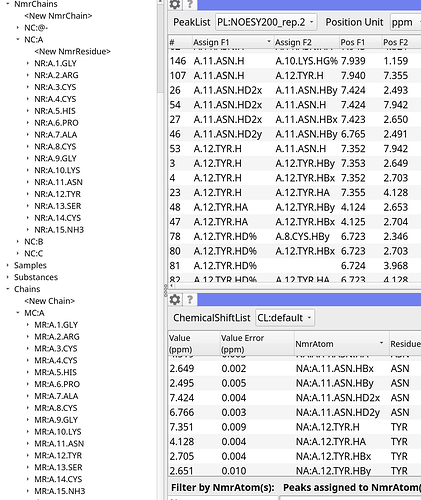
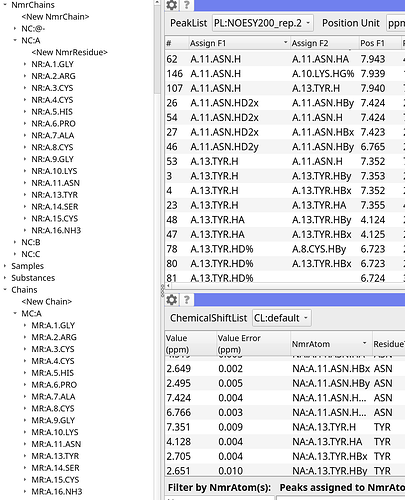
Hi! This is my chain as created from the imported sequence.
The residue renumbering in that thread appears to be for the entire chain from my understanding, such as in the case when residue one is now 500.
In my case, my chain is fine for the first half of the sequence, but due to a deletion in the middle of my sequence, everything after is shifted. I am unsure how the numbering offset would work in this scenario.
Is there a way to copy the assignments from chain A into chain B?
Hi
You can use the start and stop parameters to those methods to renumber just a part of the Chain and NmrChain which should fix your issue.
Hi,
The question is if you need your peaks to be re-assigned or do you just need chemical shifts with correct numbering?
If you need peaks, I do think it will be quite complicated to fix (should be possible), if you only need chemical shifts (so essentially nmrChain, and chain) then some handling of exported NEF file is the easiest (yet not the most elegant!) way of doing it.
Eliza
So if for example you want to insert one residue (e.g. between residue 11 and 12)…
myChain=project.chains[0]
myNmrChain = myChain.nmrChain
myChain.renumberResidues(1, 12)
myNmrChain.renumberNmrResidues(1,12)
All existing peak assignments update to follow the NmrChain changes.
Then create a new Chain with the correct sequence, delete the old one and change the new Chain’s name to A
Another way round might be to fix the NmrChain first (it will be out of sync with the Chain, but that should not be a problem) and then change the NmrChain’s name to ‘B’ or whatever to match your correct new Chain.
I think the key understanding is that Chain and NmrChain objects are more or less independent. When the assignment of an NmrAtom in an NmrResidue in an NmrChain matches a corresponding Atom, Residue and Chain then it’s “fully assigned”. Otherwise you can call the Nmr* versions what you like, although it’s probably not good for your brain to have apparently contradictory assignments hanging around!
Peak assignments are the the NMR* objects - if you change the name of the object, the peak assignment changes too.
well you could write a macro to renumber things inside ccpn as discussed by Brian
Alternatively if you want to try it in nef I have some very preliminary nef-pipelines commands that will do this
So something like
nef chains renumber —in my_file.nef | nef chains delete A:5-10 | nef chains renumber - ranges A:11 A -5
Should work
If you are interested in playing with it this way I can release this later today
[
](https://github.com/varioustoxins/NEF-Pipelines)
regards
Gary
Dr Gary S Thompson NMR Facility Manager
CCPN CoI & Working Group Member
Wellcome Trust Biomolecular NMR Facility
School of Natural Sciences
University of Kent, Canterbury, Kent, England, CT2 7NZ
 :01227 82 7117
:01227 82 7117
 : g.s.thompson@kent.ac.uk
: g.s.thompson@kent.ac.uk
orchid: ORCID
Hello!
Thank you everyone for all the suggestions! I used @bosmith 's way of solving this issue and wow, it was so much easier than I expected it to be!
I had thought that the renumbering using that line of code would apply to the whole chain, but nope! Works great, thank you again!
1 Like