Hi.
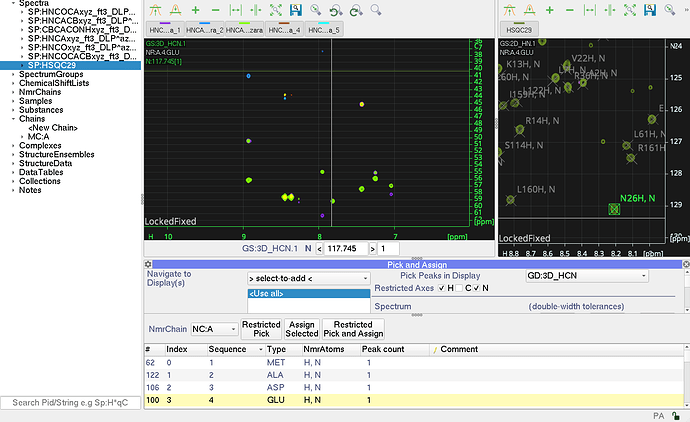
So I am trying to assign a protein bound to a ligand. I imported the 3d spectra and the hsqc. I set up my nmr chain with the sequence and then clicked set up nmr chains using my peaklist from my bound hsqc that was already assigned so I can assign the backbone carbons. However as you can see from the screen, when I click on a residue in the pick and assign module it does not navigate to that residue on either the hsqc or the 3d spectra. If I create a new peaklist with unassigned peaks then set up nmr residues it will navigate through but I want to use my already assigned hsqc peaklist. Screenshot should help.
Any help is greatly appreciated.
Thanks,
Joe
Hi Joe,
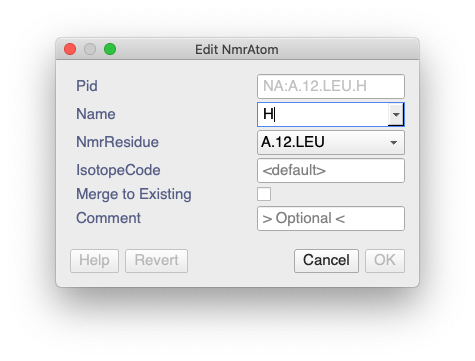
did you import the assigned peakList? I found that there seems to be a bug which means that the NmrAtoms are not assigned an isotopeCode during NEF import. This stops the navigation from working in the Pick & Assign module. You can check whether an NmrAtom has an isotopeCode by double-clicking on it in the sidebar. If the isotopeCode is simply , then it hasn’t been set.
Setting the isotopeCode isn’t straight forward and you’ll want this automated to do all in one go anyway. So have a go with this macro:
pl = get('PL:hsqc.2')
for pk in pl.peaks:
for axcde,nas in zip(pk.spectrum.axisCodes, pk.assignmentsByDimensions):
# print(nas)
pk.assignDimension(axcde, nas)
Just make sure you have selected the correct peakList at the top.
Info on running macros can be found at Running Macros
Vicky
Thanks yeah all working now on my end. I did import the peaklist as a NEF file and yes the isotopeCode missing was the cause.
Thanks a lot.
1 Like